Which problems Scientist has that Guix solves
computational reproducibility with GNU Guix
Note: The initial material had been presented to the monthly iPOP-UP “bioinfo” seminar from Univ. Paris. The slides of the presentation can be fetched here. It is joint effort with PhD student Nicolas Vallet, MD. The templates of these slides are borrowed from Ludovic Courtès’ materials with their courtesy. All opinions presented here are mine.
Reproducibility, replication crisis, Open Science (UNESCO), Ouvrir la Science, etc. are hot topics these days. They raise so many questions that it is impossible to address all of them in only one go. Here, the aim is to expose only a corner – already vast and complicated enough for keeping busy people behind the initiative GuixHPC (Guix in scientific context). Let talk about computational reproducibility!
This post is articulated in two parts. Firstly, it sets the scene by providing a pedestrian introduction to the core problem about computational reproducibility. Then secondly, it presents how to (partially) resolve …wait for it… using the tool GNU Guix.
It is a long read; although the slot of the initial presentation was 30min. Hence, it is self-contained and more you read, more you dive into details. If you are not able to schedule enough time, the first section explains the (opinionated) key points to have in mind when speaking about “Reproducible Science”1. The second section introduces technical details – necessary requirements to deeply understand why computational reproducibility is hard to achieve. Based on that, the third section asks what does it mean « I use this tool at that version »? All these three sections compose the first part. After that, I hope you will be hooked and eager to read the remaining sections introducing Guix on concrete examples – the second part.
The presentation follows a different structure but at some point, the keys are also exposed in Reproducible computations with Guix or Les enjeux et défis de la recherche reproductible (video); they are worth material.
1 What is Science?
An answer to this question means a criteria for the demarcation problem. Well, I am not convinced we can do better than Aristotle, Popper, Kuhn, Feyerabend or many more other authors. Let consider Science as the broad usual meaning. However, let assume the consensus that:
- Science requires transparency,
- a scientific result is an experiment and a numerical processing.
Based on this consensus, Science at the modern age becomes:
- Open Article: HAL, BioArxiv, ReScience and many more,
- Open Data: 1000Genomes, IMGT-HLA, Zenodo and many more,
- Open Source: BioConductor, machine learning PyTorch, long-term Software Heritage and many more.
The underlying topic barely discussed is: how to glue all that? All these items require a computational environment. We all need a computer and many programs running on it to read or write article, to fetch and analyse data, to apply processing using free software to these data, and so on. Science at the modern age requires another fourth item:
- Computational environment: that the topic we want to address here.
1.1 What is a scientific result?
Let draw a general picture separating what it concretely means a scientific result. Despite it is an obvious approximation of the reality, it helps to catch which parts are actionable when speaking about doing a scientific result.
| audit | opaque | depend? | ||||
|---|---|---|---|---|---|---|
| result | \(\longleftarrow\) | paper | + | data | + | analysis |
| data | \(\longleftarrow\) | protocol | + | instruments | + | materials |
| analysis | \(\longleftarrow\) | script | + | data | + | environment |
The table reads: to do the result, it requires to have at hand all the three, the paper describing it, and, the associated data, and, using this analysis. Another instance, to do the analysis, it requires to have at hand all the three, the script describing it, and, the associated data, and, using this environment.
For each column, the situation is at best:
- audit is the “easy” part – people describe with more or less accuracy what they are doing,
- opaque is the hard part – some events happens every once in a while (astronomy or genetic mutation), some events are highly noisy (cells in Petri dish), some instruments are protected by patents (microscope, DNA sequencer), or even some instruments are too expensive to have two (LHC, supercomputer), etc.
- depend? is the column where we can act on! This column only depends on the practise we collectively agree on. Therefore, I am advocating to evacuate that as most as possible by turning it as audit.
The boundary between data (experiment) and analysis (numerical processing) is not clearly delimited and instead it is often a grey area. How to consider trained machine learning models? Especially if they run on GPU2. Is it data or analysis? How to consider meteorological computations run after several days (week?) on supercomputer? Is it data or analysis? It is easy to find many more examples.
The true question, when speaking about producing a scientific result, is the control of the variability3. Is the variability intrinsic to the object under study? Or is it extrinsic? In this former case, what is the frame to control it? The aim in setting such frame is twofold:
- a) verify and thus convince other peers that the arguments hold and,
- b) do not build on sand, i.e., deeply understand, e.g., from where the resulting effects come.
These two items strongly require the capacity to scrutinize all the components: paper, protocol, script and more, data, analysis and… environment. That’s what Open Article, Open Data, Open Data cover for the former. For the latter, how to deal with computational environment in this scientific context?
A consequence of these two items is the capacity to redo previous results. Being able to redo what had been done is at the heart of Science and, somehow, this redo and its introspection is the core principle which turns a belief into a scientific knowledge. Let define weakly reproducibility by being able to redo what had been done. Alice runs thing at time \(t\) and Bob tries to run again thing at any another time \(t'\) possibly elsewhere. The correctness of thing and then considering if redo-thing is identical or not is the very aim of any scientific debates4. Science makes progress by arguing, thus how to argue if it is not possible to complete redo? Is it a sane discussion if computational environment is not controlled?
Still not convinced that computational environment matters for Science? Read on.
1.2 Science is collective
Any scientific practitioner knows that producing scientific knowledge is a collective activity. Many people or even teams are involved over the time. Concretely, these situations are what happens daily in the labs all around the world:
- Alice uses numerical tools R@4.1.1, FlowCore@2.4 and CATALYST@1.16.2;
- Carole collaborates on the project with Alice… but in the same time, Carole works on another project requiring the numerical tools R@4.0.4, FlowCore@2.0;
- Bob runs the same versions as Alice… but does not get the same outputs;
- Charlie upgrades their system then all is broken, reinstall everything… but is never able to restore the environment which worked;
- Dan tries to redo the Alice’s analysis months (year?) later… but hits the dependency hell5.
(Replace the tools R, FlowCore, CATALYST by any other tools from any scientific stack. Here, R is picked as example because it is very (too much?) popular in data processing from biological context.)
Any person who has seriously invested effort in numerical processing has hit at least one similar situation as Carole or Bob or Charlie or Dan. If not, please drop me an email exposing your workflow and habits.
All the situations have pragmatic solutions,
- package manager as apt or yum or etc.
- virtual environment as conda or modulefiles or etc.
- container as Docker or Singularity or etc.
and, for what my experience is worth, I have daily dealt with many of these solutions then they fail at one corner or the other – when speaking about computational reproducibility over the time. Their main drawback is that they do not tackle by design both the core issues of capturing the computational environment and being able to provide the same binaries over the time. Guix does both by design6.
2 Why is it complicated?
Let answer by another question: what is a computation? Alice runs daily R scripts to analyse complex data but does she know how this trivial computation works? (see meaning of command-line7)
alice@laptop$ R -e '1+2'
> 1+2
[1] 3
What is the issue? At first sight, one could argue that R is an Open Source
piece of software. For instance, give a look at the program entry point: it
is a C program and all is transparent. Is it really that simple?
2.1 Forensic exercise
Let dive into forensic exercise. First thing first, run a software means
that somewhere a binary program must be involved. Is this R a binary
program? The command which allows to know which software is used exactly –
where the real software is located in the system. The command file
determines the file type, i.e., if the file is a binary or text.
alice@laptop$ which R /usr/bin/R alice@laptop$ file /usr/bin/R /usr/bin/R: Bourne-Again shell script, ASCII text executable
Interesting, typing R runs a script (text executable). After studying it a
bit this script, we know two things:
- at the top, it reads
#!/bin/bash, - this
/usr/lib/R/bin/exec/Ris indeed run.
Typing R implies two binaries: the one (/bin/bash) to launch the script
named /usr/bin/R and this script calls the other binary
(/usr/lib/R/bin/exec/R). For the sake of the presentation, we let aside
the former. However, keep in mind that all what we will say to the latter
applies to both. Therefore, perhaps a first clue why it is
complicated8.
alice@laptop$ file /bin/bash /bin/bash: ELF 64-bit LSB shared object, x86-64, version 1 (SYSV), dynamically linked, interpreter /lib64/ld-linux-x86-64.so.2, for GNU/Linux 3.2.0, BuildID[sha1]=12f73d7a8e226c663034529c8dd20efec22dde54, stripped alice@laptop$ file /usr/lib/R/bin/exec/R /usr/lib/R/bin/exec/R: ELF 64-bit LSB shared object, x86-64, version 1 (SYSV), dynamically linked, interpreter /lib64/ld-linux-x86-64.so.2, for GNU/Linux 3.2.0, BuildID[sha1]=d875b0693e51ba0b8ac28ea332eb69b5ca6071af, stripped
Wow, what does ELF and dynamically linked mean?
2.2 An ELF is…
…a mythological creature – ELF is for Executable and Linkable Format. In
short, it is the binary we are looking for. Wait, we told that R is from
a C program. How do we go from this human readable C program to that
binary.
Recipe to make Yogurt: take some Milk and mix to some Skyr9 (other kind of Yogurt). This analogy applies to make binary and reads,
| Yogurt | \(\longleftarrow\) | Milk | + | Skyr |
| binary | source | binary | ||
/usr/lib/R/bin/exec/R |
main.c |
C compiler |
It is easy to imagine that the C compiler is an executable too. It
comes from an human-readable source code and another executable. This
other executable is also from an human-readable source code and
another-bis executable. Recursion, oh recursion! At the end, a chicken
or the egg problem arises (named bootstrapping compiler). Curious reader
might be interested by this entry point.
It is a bit oversimplified since the file main.c alone is obviously not
enough to produce all the features that R provides. Instead, all the
source files are first transformed as binary object and then second linked
altogether to produce the resulting unique binary.
In summary, the key component to keep in mind is that building a binary program requires one or more other binary program. It is often named build-time dependencies.
2.3 Dynamically linked means
Let examine /usr/lib/R/bin/exec/R by using the command-line tool ldd
which prints the shared object dependencies.
alice@laptop$ ldd /usr/lib/R/bin/exec/R libblas.so.3 => /usr/lib/x86_64-linux-gnu/libblas.so.3 libR.so => /usr/lib/libR.so libgfortran.so.5 => /usr/lib/x86_64-linux-gnu/libgfortran.so.5 libquadmath.so.0 => /usr/lib/x86_64-linux-gnu/libquadmath.so.0 libblas.so.3 => /usr/lib/x86_64-linux-gnu/libblas.so.3 libR.so => /usr/lib/libR.so libgfortran.so.5 => /usr/lib/x86_64-linux-gnu/libgfortran.so.5 libquadmath.so.0 => /usr/lib/x86_64-linux-gnu/libquadmath.so.0 ... many more ...
Euh, what does it mean? It means that the binary /usr/lib/R/bin/exec/R
is dynamically linked to all these software. And wait for it, there are
binaries too. All we said in the previous subsection apply for each
item. We are explaining why it is complicated, so let slash at that!
Let pick one: BLAS provides all the basic linear algebra subprograms. The
command ls -l allows to list all the properties of a file. On Ubuntu
18.04, it reads,
alice@laptop$ ls -l /usr/lib/x86_64-linux-gnu/libblas.so.3 lrwxrwxrwx 1 root root 47 avril 21 11:44 /usr/lib/x86_64-linux-gnu/libblas.so.3 -> /etc/alternatives/libblas.so.3-x86_64-linux-gnu alice@laptop$ ls -l /etc/alternatives/libblas.so.3-x86_64-linux-gnu lrwxrwxrwx 1 root root 43 avril 21 11:44 /etc/alternatives/libblas.so.3-x86_64-linux-gnu -> /usr/lib/x86_64-linux-gnu/blas/libblas.so.3 alice@laptop$ ls -l /usr/lib/x86_64-linux-gnu/blas/libblas.so.3 lrwxrwxrwx 1 root root 16 nov. 28 2017 /usr/lib/x86_64-linux-gnu/blas/libblas.so.3 -> libblas.so.3.7.1 alice@laptop$ file /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.7.1 /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.7.1: ELF 64-bit LSB shared object, x86-64, version 1 (SYSV), dynamically linked, BuildID[sha1]=ca15f6c1fdcbd9438bb94298f38bf7fa0e99bc5f, stripped
The file /usr/lib/x86_64-linux-gnu/libblas.so.3 is a symbolic file which
points to the file /etc/alternatives/libblas.so.3-x86_64-linux-gnu which
is another symbolic link /usr/lib/x86_64-linux-gnu/blas/libblas.so.3
which points to the binary
/usr/lib/x86_64-linux-gnu/blas/libblas.so.3.7.1.
In summary, the key component to keep in mind is that running a binary program requires one or more other binary programs. It is often named run-time dependencies.
We got it, it is complicated. The solution is to capture the graph of
dependencies and package managers (apt, yum, conda, brew, etc.)
does already that job. Indeed, but is it that simple?
3 What is a version?
Again a story about producing Yogurt. As we said previously, the recipe to procude binary is similar to the recipe to produce Yogurt, it reads,
| Yogurt | \(\longleftarrow\) | Milk | + | Skyr | + | Utensils |
| binary | source | binary | links | |||
/usr/lib/R/bin/exec/R |
main.c |
C compiler |
Deps. (libblas, etc.) |
Using this frame, when Alice says I use R@4.1.1 for the analysis, it implicitly means,
| R@4.1.1 | \(\longleftarrow\) | main.c @4.1.1 |
+ | executable @A | + | links @C |
What happens if Bob uses another versions for executable@B and link@D? Can the end binary result R@4.1.1 be considered as the same version? Or is it another version?
For instance, Alice says she use R@4.1.1, thus she somehow runs,
alice@laptop$ install r@4.1.1
alice@laptop$ R --version
R version 4.1.1 (2021-08-10) -- "Kick Things"
and Bob runs this very same version R@4.1.1 but on another machine,
bob@cluster$ install r@4.1.1
bob@cluster$ R --version
R version 4.1.1 (2021-08-10) -- "Kick Things"
then Bob does not get the same result, e.g., precision or other. Why is it
different? Both use the same versions after all. Because Bob knows Alice,
he asks her the binary /usr/lib/R/bin/exec/R to diff against his own.
Result: Binary files differ. It is the same source version but not the
same binary version.
We claim that I use R@4.1.1 for the analysis is not enough. Such information only provides the version of the source. Then, too many combinations are possible for the build-time (executable) and run-time (links) dependencies to get the same environment on different machines.
When something is going wrong, it is hard, nor impossible, to say from where it comes. Is it because the computational environment is different? Is it because the analysis has flaws? Is it something else?
In summary, the key component to keep in mind is tools are required to capture the environment ( @4.1.1, @A, @C)
Computional reproducibility, which just means redo later and elsewhere the same exact analysis, requires more than only a list of package@version. It requires to capture how it is built. For example, R@4.1.1 requires 400+ other packages to build and run. This is what Guix does: it captures the complete graph of dependencies.
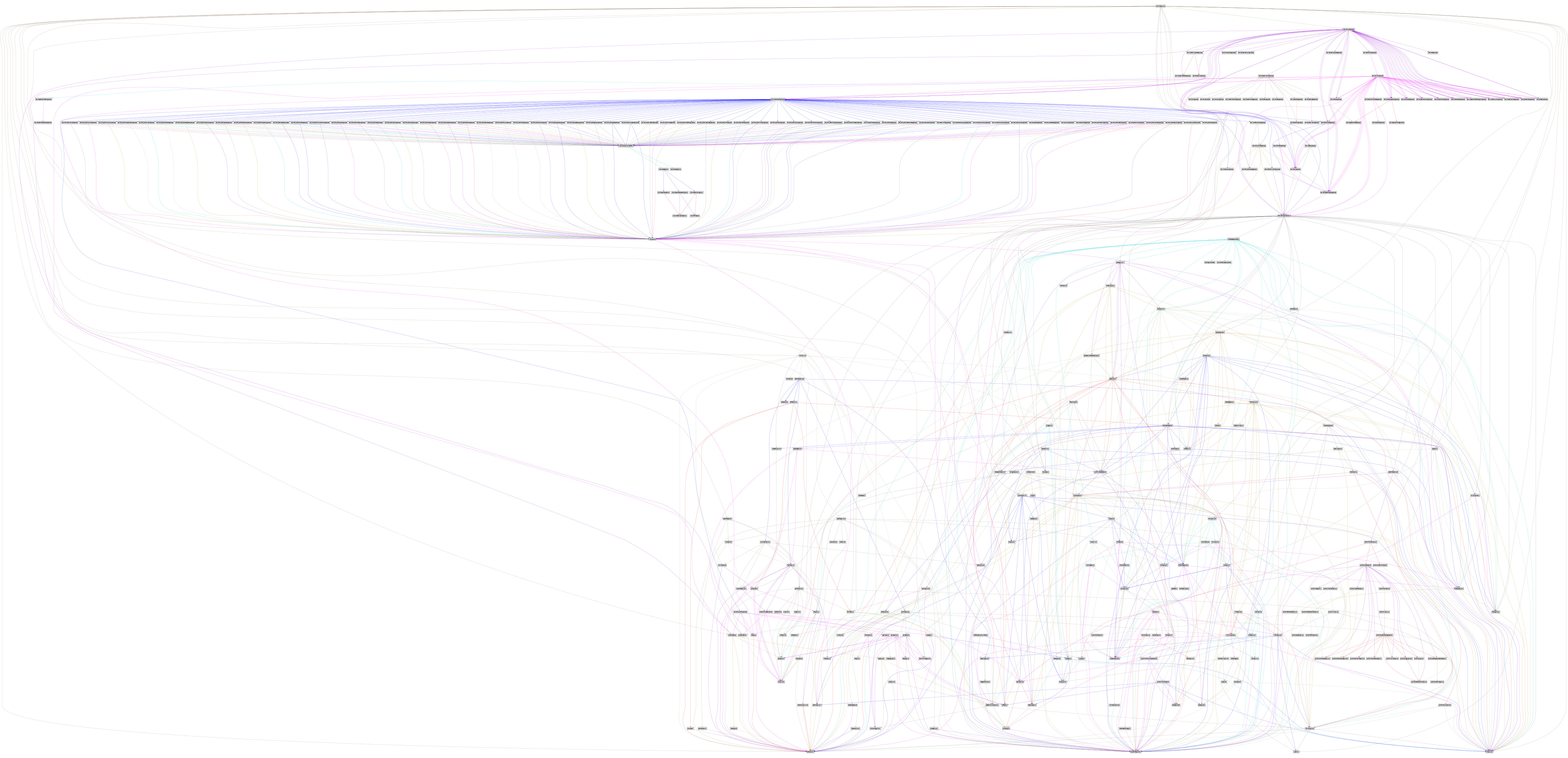
guix graph r-minimalThe second part will explain concrete solutions for the working scientist.
Join the fun, join Guix for Science!
Footnotes:
Reproducible Science, probably a logical tautology.
Programming on GPU requires a lot of opaque binary blobs.
Variability in the statistical meaning.
See all the scientific controversies.
Hit dependencies hell is at best!
Functional package management pioneered by Nix.
alice@laptop$ means
Alice executes on her laptop the command-line which follows the symbol
$.
And consider too that another script programming the analysis and using many other binaries (packages) is also involved.
Skyr, Icelandic strained yogurt, delicious!
